🤔 NumPy & Pandas
NumPy와 Pandas는 머신러닝을 공부한다면 절대 빠질 수 없는 필수 라이브러리입니다.
많은 사람들이 Numpy와 Pandas에 대한 설명을 해두었고 앞으로 공부할 내용에서도 많이 사용할 것이므로 간단하게 코드로 설명하고 넘어가겠습니다.
🔎 NumPy
NumPy는 수학적인 연산 및 다차원 배열 (Array, Matrix, Tensor)의 연산을 가능하게 해주는 Python의 Library입니다.
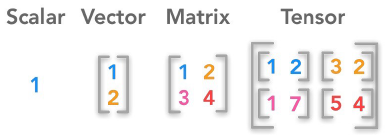
아래는 NumPy를 사용한 예시입니다.
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
import numpy as np
python_list = [1, 2, 3, 4, 5]
python_list, type(python_list)
([1, 2, 3, 4, 5], list)
numpy_array = np.array([1, 2, 3, 4, 5])
numpy_array, type(numpy_array)
(array([1, 2, 3, 4, 5]), numpy.ndarray)
numpy_matrix = np.array([[0 for _ in range(4)] for _ in range(3)])
print(numpy_matrix.shape)
numpy_matrix
(3, 4)
array([[0, 0, 0, 0],
[0, 0, 0, 0],
[0, 0, 0, 0]])
numpy_zeros = np.zeros((3, 4))
print(numpy_zeros.shape)
numpy_zeros
(3, 4)
array([[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.]])
numpy_tensor = np.array(
[
[
[
i for i in range(3)
] for _ in range(3)
] for _ in range(5)
]
)
for i, v in enumerate(numpy_tensor):
print(i, v.shape)
print('numpy_tensor의 shape : ', numpy_tensor.shape)
0 (3, 3)
1 (3, 3)
2 (3, 3)
3 (3, 3)
4 (3, 3)
numpy_tensor의 shape : (5, 3, 3)
np_1 = np.array([1, 2, 3])
np_2 = np.array([3, 3, 3])
print(np.add(np_1, np_2))
print(np.subtract(np_2, np_1))
print(np.multiply(np_2, np_1))
print(np.divide(np_2, np_1))
[4 5 6]
[2 1 0]
[3 6 9]
[3. 1.5 1. ]
product_1 = np.array([[1, 0], [0, 1]])
product_2 = np.array([[1, 2], [3, 4]])
print(np.matmul(product_1, product_2))
[[1 2]
[3 4]]
product_3 = np.array([[1, 2, 3], [3, 2], [1, 1]])
product_3
/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:1: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
"""Entry point for launching an IPython kernel.
array([list([1, 2, 3]), list([3, 2]), list([1, 1])], dtype=object)
product_3 = np.array([[1, 2], [3, 2], [1, 1]])
product_3
array([[1, 2],
[3, 2],
[1, 1]])
행렬 곱에 있어서는 shape를 꼭 맞춰줘야 한다는 예시입니다.
np.matmul(product_1, product_3)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-23-69fb13202340> in <module>
----> 1 np.matmul(product_1, product_3)
ValueError: matmul: Input operand 1 has a mismatch in its core dimension 0, with gufunc signature (n?,k),(k,m?)->(n?,m?) (size 3 is different from 2)
product_1 @ product_3.reshape(2, 3)
array([[1, 2, 3],
[2, 1, 1]])
product_1 @ product_3.transpose()
array([[1, 3, 1],
[2, 2, 1]])
np_concat = np.concatenate((np_1, np_2))
print(np_concat, np_concat.shape)
np_transpose = np.reshape(np_concat, (2, 3))
print(np_transpose.shape)
np_t = np_transpose.transpose()
print(np_t.shape)
[1 2 3 3 3 3] (6,)
(2, 3)
(3, 2)
🔎 Pandas
Pandas는 데이터 분석에 사용되는 도구이며 정형데이터를 처리하는 가장 대표적인 도구입니다.
csv파일을 아래와 같은 형식으로 사용자에게 보여줍니다.
NumPy와 마찬가지로 간단하게 코드를 통해 살펴보겠습니다.
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
import pandas as pd
import numpy as np
data = pd.read_csv('/content/drive/MyDrive/2022/CNU/ML/Iris.csv')
data
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
| ... | ... | ... | ... | ... | ... |
| 146 | 6.7 | 3.0 | 5.2 | 2.3 | Iris-virginica |
| 147 | 6.3 | 2.5 | 5.0 | 1.9 | Iris-virginica |
| 148 | 6.5 | 3.0 | 5.2 | 2.0 | Iris-virginica |
| 149 | 6.2 | 3.4 | 5.4 | 2.3 | Iris-virginica |
| 150 | 5.9 | 3.0 | 5.1 | 1.8 | Iris-virginica |
150 rows × 6 columns
data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Id 150 non-null int64
1 SepalLengthCm 150 non-null float64
2 SepalWidthCm 150 non-null float64
3 PetalLengthCm 150 non-null float64
4 PetalWidthCm 150 non-null float64
5 Species 150 non-null object
dtypes: float64(4), int64(1), object(1)
memory usage: 7.2+ KB
data.head()
data.tail()
| 146 | 6.7 | 3.0 | 5.2 | 2.3 | Iris-virginica |
| 147 | 6.3 | 2.5 | 5.0 | 1.9 | Iris-virginica |
| 148 | 6.5 | 3.0 | 5.2 | 2.0 | Iris-virginica |
| 149 | 6.2 | 3.4 | 5.4 | 2.3 | Iris-virginica |
| 150 | 5.9 | 3.0 | 5.1 | 1.8 | Iris-virginica |
del data['Id']
data.head()
| 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
data.mean(), data.std()
/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:1: FutureWarning: Dropping of nuisance columns in DataFrame reductions (with 'numeric_only=None') is deprecated; in a future version this will raise TypeError. Select only valid columns before calling the reduction.
"""Entry point for launching an IPython kernel.
(SepalLengthCm 5.843333
SepalWidthCm 3.054000
PetalLengthCm 3.758667
PetalWidthCm 1.198667
dtype: float64, SepalLengthCm 0.828066
SepalWidthCm 0.433594
PetalLengthCm 1.764420
PetalWidthCm 0.763161
dtype: float64)
data.describe()
| 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| 5.843333 | 3.054000 | 3.758667 | 1.198667 |
| 0.828066 | 0.433594 | 1.764420 | 0.763161 |
| 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| 7.900000 | 4.400000 | 6.900000 | 2.500000 |
data.loc[:3, :"SepalWidthCm"]
| 5.1 | 3.5 |
| 4.9 | 3.0 |
| 4.7 | 3.2 |
| 4.6 | 3.1 |
data.iloc[:3, :-1]
| 5.1 | 3.5 | 1.4 | 0.2 |
| 4.9 | 3.0 | 1.4 | 0.2 |
| 4.7 | 3.2 | 1.3 | 0.2 |
normal_data = (data.iloc[:, : -1] - data.iloc[:, :-1].mean()) / data.iloc[:, :-1].std()
normal_data
| -0.897674 | 1.028611 | -1.336794 | -1.308593 |
| -1.139200 | -0.124540 | -1.336794 | -1.308593 |
| -1.380727 | 0.336720 | -1.393470 | -1.308593 |
| -1.501490 | 0.106090 | -1.280118 | -1.308593 |
| -1.018437 | 1.259242 | -1.336794 | -1.308593 |
| ... | ... | ... | ... |
| 1.034539 | -0.124540 | 0.816888 | 1.443121 |
| 0.551486 | -1.277692 | 0.703536 | 0.918985 |
| 0.793012 | -0.124540 | 0.816888 | 1.050019 |
| 0.430722 | 0.797981 | 0.930239 | 1.443121 |
| 0.068433 | -0.124540 | 0.760212 | 0.787951 |
150 rows × 4 columns
index = np.where(data['SepalLengthCm'] > 6.0)
index
(array([ 50, 51, 52, 54, 56, 58, 63, 65, 68, 71, 72, 73, 74,
75, 76, 77, 86, 87, 91, 97, 100, 102, 103, 104, 105, 107,
108, 109, 110, 111, 112, 115, 116, 117, 118, 120, 122, 123, 124,
125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137,
139, 140, 141, 143, 144, 145, 146, 147, 148]),)
data.Species.unique()
array(['Iris-setosa', 'Iris-versicolor', 'Iris-virginica'], dtype=object)
data.Species.value_counts()
Iris-setosa 50
Iris-versicolor 50
Iris-virginica 50
Name: Species, dtype: int64
target_dict = {}
for i, v in enumerate(data.Species.unique()):
target_dict[i] = v
target_dict
{0: 'Iris-setosa', 1: 'Iris-versicolor', 2: 'Iris-virginica'}
label_encoded_species = pd.Series([None for i in range(data.shape[0])])
for i in range(len(target_dict)):
label_encoded_species[data['Species'] == target_dict[i]] = i
print(data['Species'][0], label_encoded_species[0])
print(data['Species'][50], label_encoded_species[50])
print(data['Species'][100], label_encoded_species[100])
Iris-setosa 0
Iris-versicolor 1
Iris-virginica 2
from sklearn.preprocessing import LabelEncoder
label_encoded_series_sklearn = LabelEncoder().fit_transform(data[['Species']])
label_encoded_series_sklearn
/usr/local/lib/python3.7/dist-packages/sklearn/preprocessing/_label.py:115: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().
y = column_or_1d(y, warn=True)
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2])
from sklearn.preprocessing import OneHotEncoder
df = OneHotEncoder(sparse=False).fit_transform(data[['Species']])
print(data['Species'][0], df[0])
print(data['Species'][50], df[50])
print(data['Species'][100], df[100])
Iris-setosa [1. 0. 0.]
Iris-versicolor [0. 1. 0.]
Iris-virginica [0. 0. 1.]
label = pd.DataFrame(df, columns=data.Species.unique(), dtype=int)
label
| 1 | 0 | 0 |
| 1 | 0 | 0 |
| 1 | 0 | 0 |
| 1 | 0 | 0 |
| 1 | 0 | 0 |
| ... | ... | ... |
| 0 | 0 | 1 |
| 0 | 0 | 1 |
| 0 | 0 | 1 |
| 0 | 0 | 1 |
| 0 | 0 | 1 |
150 rows × 3 columns
data = pd.concat([data.iloc[:, :-1], label], axis=1)
data
| 5.1 | 3.5 | 1.4 | 0.2 | 1 | 0 | 0 |
| 4.9 | 3.0 | 1.4 | 0.2 | 1 | 0 | 0 |
| 4.7 | 3.2 | 1.3 | 0.2 | 1 | 0 | 0 |
| 4.6 | 3.1 | 1.5 | 0.2 | 1 | 0 | 0 |
| 5.0 | 3.6 | 1.4 | 0.2 | 1 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... |
| 6.7 | 3.0 | 5.2 | 2.3 | 0 | 0 | 1 |
| 6.3 | 2.5 | 5.0 | 1.9 | 0 | 0 | 1 |
| 6.5 | 3.0 | 5.2 | 2.0 | 0 | 0 | 1 |
| 6.2 | 3.4 | 5.4 | 2.3 | 0 | 0 | 1 |
| 5.9 | 3.0 | 5.1 | 1.8 | 0 | 0 | 1 |
150 rows × 7 columns
data = data.reset_index(drop=True)
data
| 5.1 | 3.5 | 1.4 | 0.2 | 1 | 0 | 0 |
| 4.9 | 3.0 | 1.4 | 0.2 | 1 | 0 | 0 |
| 4.7 | 3.2 | 1.3 | 0.2 | 1 | 0 | 0 |
| 4.6 | 3.1 | 1.5 | 0.2 | 1 | 0 | 0 |
| 5.0 | 3.6 | 1.4 | 0.2 | 1 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... |
| 6.7 | 3.0 | 5.2 | 2.3 | 0 | 0 | 1 |
| 6.3 | 2.5 | 5.0 | 1.9 | 0 | 0 | 1 |
| 6.5 | 3.0 | 5.2 | 2.0 | 0 | 0 | 1 |
| 6.2 | 3.4 | 5.4 | 2.3 | 0 | 0 | 1 |
| 5.9 | 3.0 | 5.1 | 1.8 | 0 | 0 | 1 |
150 rows × 7 columns
data.to_csv('new_Iris.csv', index=False)
🔎 Reference
https://numpy.org/doc/stable/user/
NumPy user guide — NumPy v1.23 Manual
numpy.org
https://numpy.org/doc/stable/reference/
NumPy Reference — NumPy v1.23 Manual
numpy.org
https://pandas.pydata.org/docs/getting_started/overview.html
Package overview — pandas 1.4.4 documentation
Package overview pandas is a Python package providing fast, flexible, and expressive data structures designed to make working with “relational” or “labeled” data both easy and intuitive. It aims to be the fundamental high-level building block for d
pandas.pydata.org
'AI > Machine Learning' 카테고리의 다른 글
| [ML] Regression(회귀)(1) - Linear Regression(선형 회귀) (0) | 2022.11.11 |
|---|---|
| [ML] 베이지안 분류기(Bayesian Classifier)(4) - Bayesian Classifier (0) | 2022.10.14 |
| [ML] 베이지안 분류기(Bayesian Classifier)(3) - Parameter Estimation (0) | 2022.10.14 |
| [ML] 베이지안 분류기(Bayesian Classifier)(2) - 베이즈 정리(Bayes' Theorem) (0) | 2022.10.14 |
| [ML] 베이지안 분류기(Bayesian Classifier)(1) - 알아보기 (0) | 2022.10.03 |
![[ML] NumPy & Pandas](https://img1.daumcdn.net/thumb/R750x0/?scode=mtistory2&fname=https%3A%2F%2Fblog.kakaocdn.net%2Fdn%2FzYvI5%2FbtrMnnbxBt3%2FnQrQ30rNOVefhj5TKKkxx0%2Fimg.jpg)